Success Stories
Aquatic eDNA macroinvertebrate monitoring of chalk streams

A collaboration between
Project Snapshot
Overview
- River Action Project Chairman Charles Watson conducted eDNA surveys of aquatic macroinvertebrates in three Hampshire chalk streams as part of the Upper Itchen Initiative.
- Despite having no previous ecological training, Charles was able to collect the eDNA samples and send them to the NatureMetrics laboratories for analysis quickly and safely.
- The project delivered data across 3 different sites and highlighted significant differences in macroinvertebrate diversity at sites with different suspected levels of pollution.

No items found.
The Challenge
The River Itchen and its tributaries are part of an important chalk stream network, and local conservation groups and charities have expressed concern about the impacts of businesses in the area. With limited resources, the group hoped to collect data on macroinvertebrate communities from across the landscape in order to characterise the ecological impacts of industrial activity on the River Itchen.

No items found.
Our Role
- Following an online training session, eDNA samples were collected spread across three separate chalk streams.
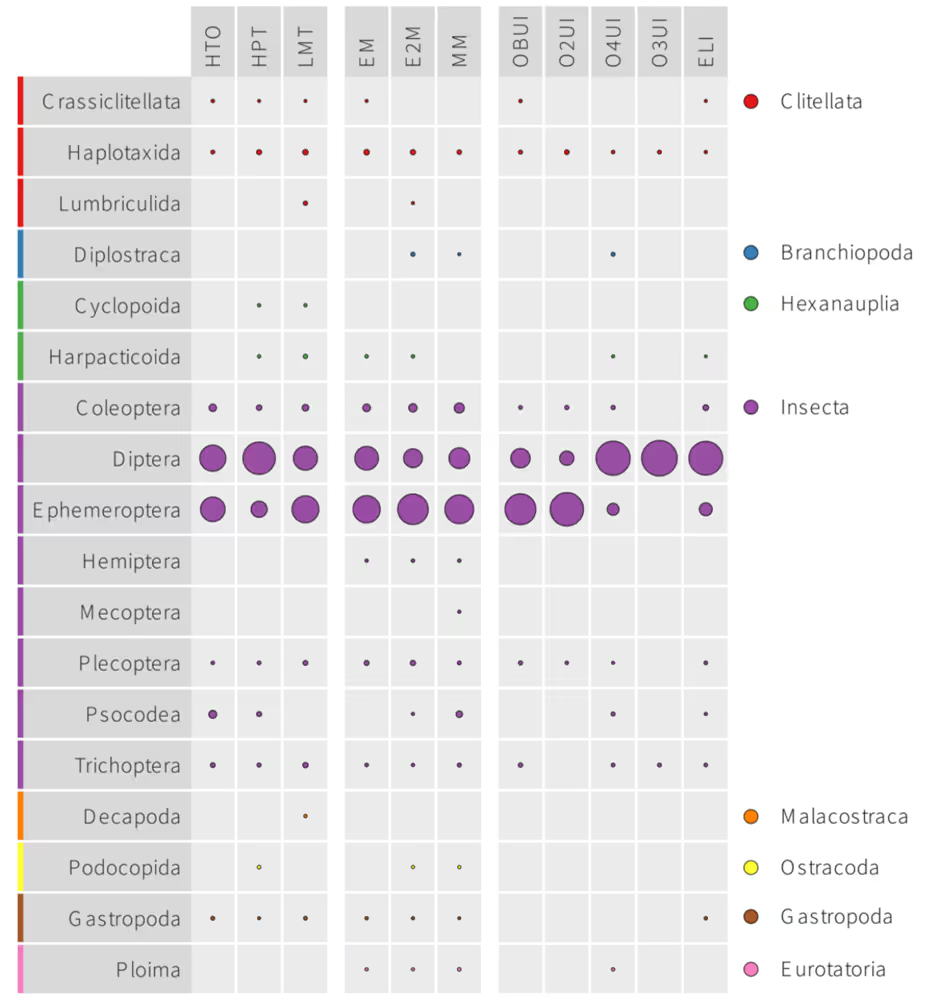
- In the lab, we analysed the samples using the eDNA Macroinvertebrate metabarcoding pipeline. This analysis specifically targets aquatic insects, including pollution-sensitive groups such as mayflies (Ephemeroptera), stoneflies (Plecoptera) and caddisflies (Trichoptera).
- The results were detailed in an easy-to-understand report that highlighted significant differences in macroinvertebrate diversity at sites with different suspected levels of pollution.

No items found.
The Findings
Highlights of the report included:
- A total of 339 taxa were detected, belonging to 17 orders, 54 families and 187 genera. This included midges (Diptera: Chironomidae) which are extremely challenging to identify with conventional morphology and are therefore often overlooked or lumped into a single taxon in surveys.
- The most abundant groups in terms of sequence read count were flies (Diptera) and mayflies (Trichoptera). Other important groups such as stoneflies (Plecoptera) and caddisflies (Trichoptera) were also detected, but the freshwater shrimp Gammarus gammarus was a notable absence from the dataset since this analysis really targets aquatic insects.
- The most abundant species in terms of sequence reads was the Large Dark Olive (Baetis rhodani).

No items found.
The Impact

No items found.

More than 600 companies in 109 countries choose NatureMetrics as their Nature Intelligence Partner
Explore more success stories
No items found.



.avif)