Soil eDNA helps Jacobs survey biodiversity for road project
Using biodiversity data from across four habitats to help to inform management decisions for woodland creation project

A collaboration between
Project Snapshot
Overview
Jacobs was contracted by Transport Scotland to undertake an impact assessment for a major road widening project. This had determined that compensatory woodland planting was required, and soil translocation had been proposed as a measure to improve the success of the new habitat creation.
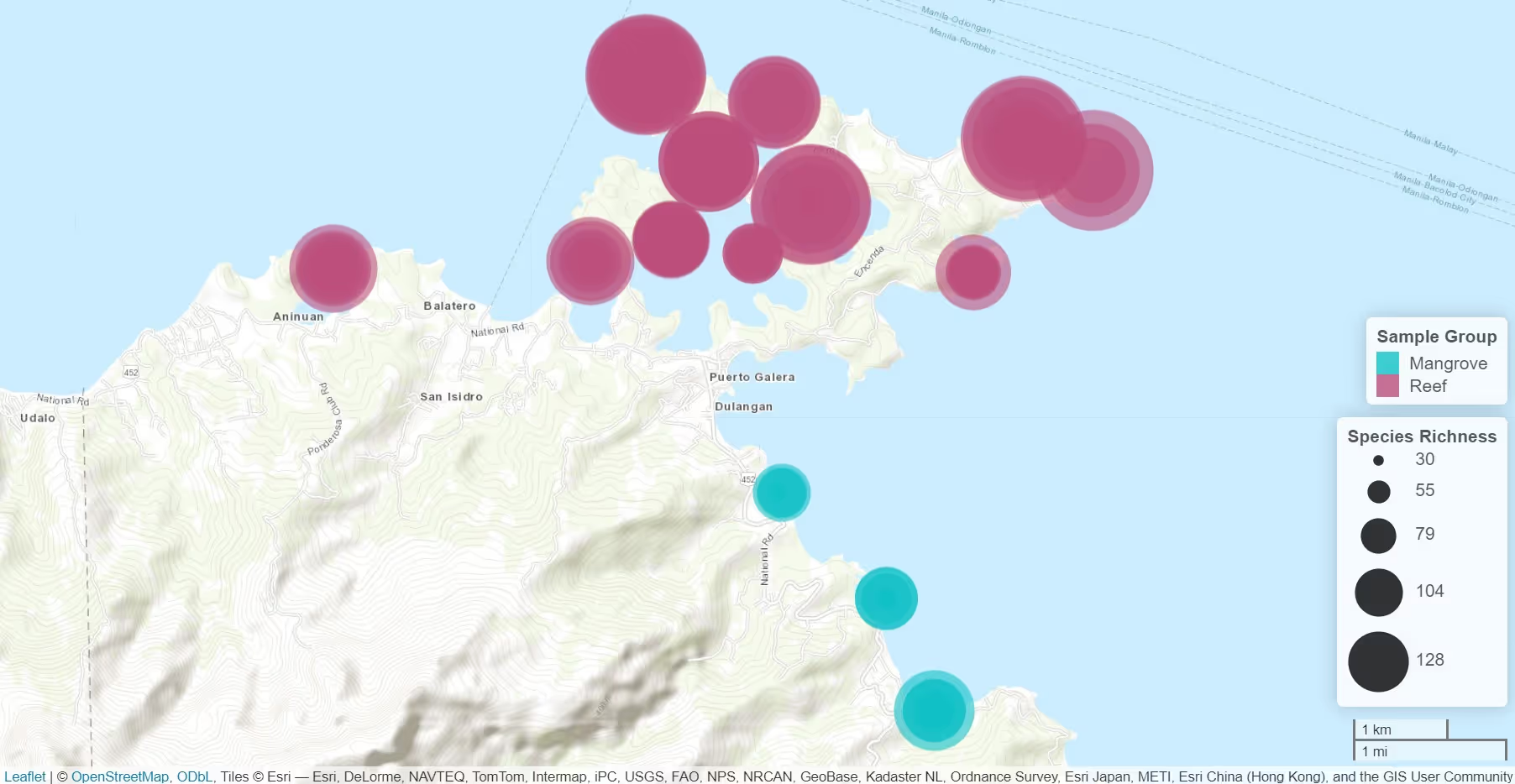
Prior to soil translocation, Jacobs used our soil environmental DNA (eDNA) service to conduct a baseline survey of soil biodiversity in four habitat types: ancient birch woodland, non-ancient birch woodland, ancient coniferous plantation woodland, and a grazing pasture grassland area designated for woodland habitat creation. Bulk mesofauna samples (where specimens had been separated from the soil and preserved in ethanol) were also analysed, but these provided less powerful data than analysis of DNA extracted directly from the soil.
Our data showed that the birch woodland, conifer woodland and grassland habitats were clearly distinguishable based on their soil biodiversity, with the grassland supporting a particularly distinct community. This data will support decision-making in selecting soils for translocation and will provide a way of evaluating the progress of the compensation planting areas towards a habitat that is ecologically equivalent to the woodland that was lost.

The Challenge
Biodiversity Baseline for Meaningful Target-Setting
Soil biodiversity is a critical component of terrestrial ecosystems and can provide a holistic picture of ecosystem type and condition.
Monitoring soil biodiversity is extremely challenging using traditional morphological taxonomy because the organisms are tiny and poorly known. Even expert taxonomists can usually only identify certain taxonomic groups, which makes it slow and expensive – if not impossible – to obtain detailed data across the whole soil community. Some groups (e.g. bacteria and some fungal groups) are not realistically feasible to comprehensively survey with traditional survey methods.
eDNA was identified as an efficient and cost-effective way of generating soil biodiversity data across multiple taxonomic groups to provide a baseline against which to monitor progress and evaluate the success of compensatory habitat creation.





Our Role
Using eDNA to Characterise Habitats
The use of DNA-based methods opens up a new frontier in understanding soil biodiversity and characterising habitats. It allows us to gain access to soil biodiversity, providing data on thousands of species across multiple taxonomic groups for comprehensive assessment and monitoring.
Our aims were to:
- Characterise the pre-impact woodland soil communities and compare these with the grassland habitat where new woodland will be created. This sets the baseline for monitoring the success of the woodland habitat creation.
- Compare the soil biodiversity of the different woodland habitats to inform the soil translocation strategy.
- Compare different taxonomic groups to determine which are most informative for differentiating habitats and tracking restoration.
- Compare DNA extraction from soil samples vs bulk mesofauna samples to determine the best method of measuring soil faunal biodiversity.

The Findings
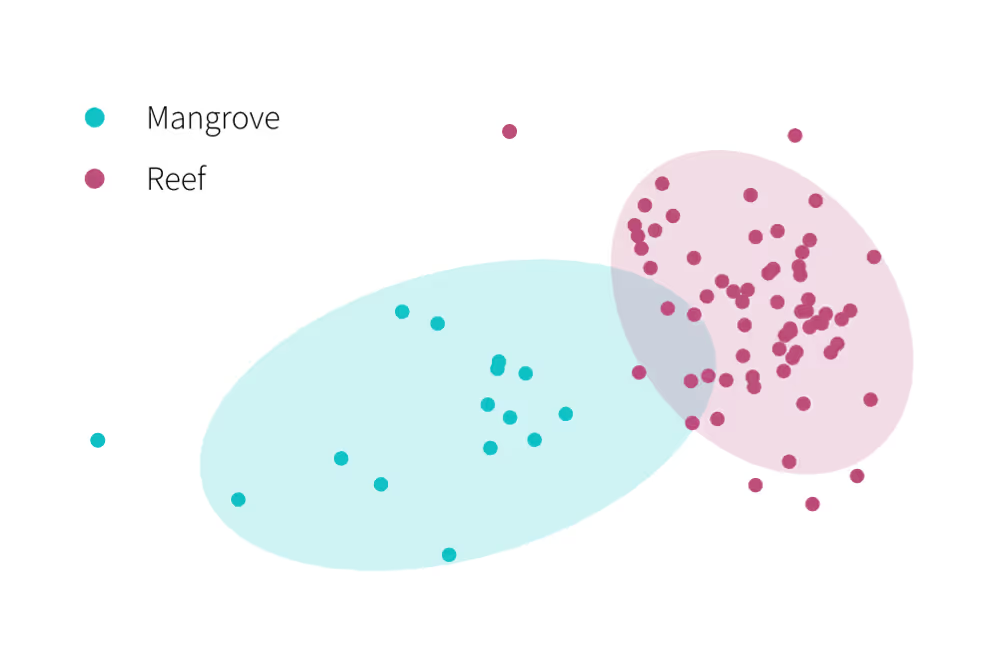
Jacobs were able to visualise the difference in soil biodiversity between grassland and woodland habitats at their site, enabling them to characterise the ‘gap’ that needs to be bridged through habitat creation.
They now have a solid baseline against which to monitor as the project progresses. It is anticipated new samples will be collected in subsequent years and added to this dataset to show how communities are changing over time and relative to the baseline and control sites.
Jacobs can now objectively compare the soil communities of the different woodland types to inform the soil translocation mitigation strategy and establish which soils are best suited to translocation.
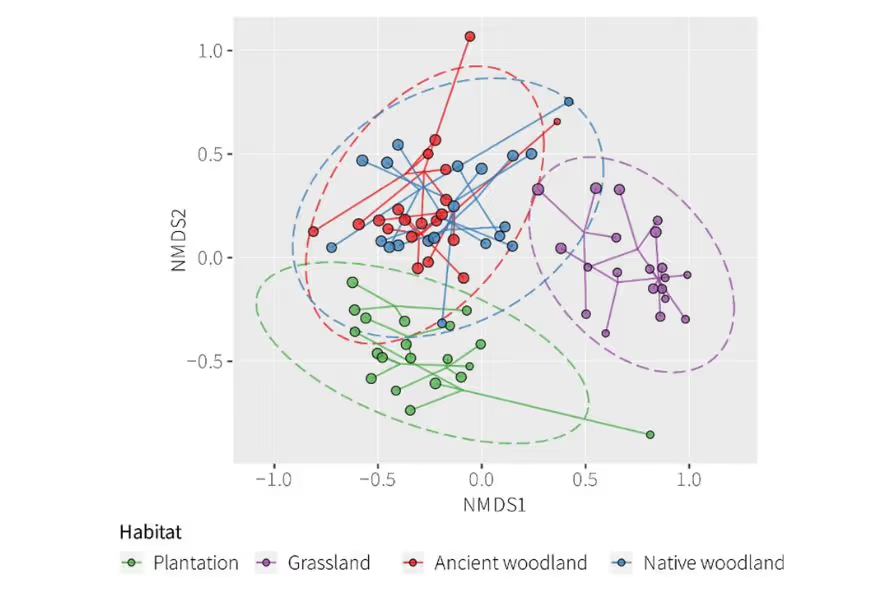
Figure 1. An ordination plot for soil faunal communities. Each point represents one sample and is coloured by habitat. Points that are closer together have more species in common than points that are further apart.
Soil eDNA provided a dataset of almost 3,000 species-level taxa.
There were clear variations in the communities associated with the different types of habitat. In particular, grassland soil communities differed significantly from those in the woodland that would likely be affected by the road scheme (Figure 1). The grassland habitat also had lower fungal and faunal richness (number of taxa) compared to the woodlands.
The soil faunal and fungal datasets independently showed very similar patterns of variation among the different habitats, which adds confidence to interpretations. Bacteria were slightly less informative for differentiating habitats but still varied significantly between the woodland and grassland habitats.
There was no significant difference between the soil communities of the ancient birch woodland and the non-ancient birch woodland. Additional sampling would be required to strengthen this finding, but it may have implications for the mitigation strategy in terms of the range of woodland soils that could be used to assist the successful establishment of new woodland compensatory planting sites.
We also carried out analysis of soil fauna that had been extracted from large 100cm3 soil cores using heat sources, as is typical for morphological analysis. This yielded only around 10% the number of species detected by direct analysis of the soil samples, with a strong bias towards arthropo (especially beetles, mites and springtails). Major groups such as nematodes were entirely missing and the power of this data to characterise and differentiate habitats was much lower. Furthermore, direct metabarcoding of the soil samples carried less risk of samples being lost through transporting between laboratories and overall was cheaper than the two-step method of pre-extraction then DNA metabarcoding.
This survey was able to set the baseline for habitat creation. As grassland areas are planted to create woodland, one of the success measures will be the extent to which the soil communities become more like those in the woodland baseline, indicating the establishment of a truly representative woodland habitat. This could be accelerated by mitigation strategies such as translocating woodland soils to grassland areas.

The Impact
A Multihabitat Baseline
Jacobs were able to visualise the difference in soil biodiversity between grassland and woodland habitats at their site, enabling them to characterise the ‘gap’ that needs to be bridged through habitat creation.
They now have a solid baseline against which to monitor as the project progresses. It is anticipated new samples will be collected in subsequent years and added to this dataset to show how communities are changing over time and relative to the baseline and control sites.
Jacobs can now objectively compare the soil communities of the different woodland types to inform the soil translocation mitigation strategy and establish which soils are best suited to translocation.







.avif)